Translate this page into:
Mutation screening of genes associated with congenital talipes equinovarus in pakistani families
2 Department of Anatomy, Institute of Basic Medical Science, Khyber Medical University, Peshawar, Pakistan
3 Department of Anatomy, College of Medicine, Al-Rayan Colleges, Taibah University, Almadinah Almunawwarah, KSA
4 Center for Genetics and Inherited Diseases, Taibah University, Almadinah Almunawwarah, KSA
Corresponding Author:
Sulman Basit
Center for Genetics and Inherited Diseases, Taibah University, 30001 Almadinah Almunawarah
KSA
sbasit.phd@gmail.com
| How to cite this article: Khan YN, Huma Z, Khan U, Basit S. Mutation screening of genes associated with congenital talipes equinovarus in pakistani families. J Musculoskelet Surg Res 2020;4:25-30 |
Abstract
Objectives: Congenital talipes equinovarus (CTEV) is characterized by midfoot cavus, forefoot adducts, hindfoot varus, and equinus. The condition leads to difficulty in walking. CTEV is one of the most common congenital birth defects and an incidence of 1–2/1000 newborns has been reported. The etiology of the CTEV is not completely understood. Multiple etiological factors, both environmental and genetic, have been implicated in the pathogenesis of CTEV. Genetic factors associated with the development of CTEV include mutations in genes related to limb patterning and development of muscle and blood vessels. The present study aims to screen four genes to detect the mutation underlying CTEV in families from Pakistan. Methods: Peripheral blood samples were collected from ten individuals from two Pakistani families with CTEV segregating in an autosomal recessive manner. Genomic DNA was isolated, followed by primer designing and Sanger sequencing of four known genes associated with the isolated form of CTEV (TBX4, PITX1, HOXD12, and HOXD9). Results: Clinical diagnoses of the affected individuals were made by consultant orthopedics. Sequencing analyses revealed that the four candidate genes screened here are not responsible for the CTEV phenotype in both families. Conclusion: Failure to detect sequence variant in four known genes associated with the isolated form of CTEV lead us to the conclusion that TBX4, PITX1, HOXD12, and HOXD9 mutations are not responsible for CTEV in both families. Therefore, genome-wide studies, including whole-genome single nucleotide polymorphism genotyping and whole-exome sequencing, are required to identify the underlying genetic defects in these families.Introduction
Skeletal disorders consist of a wide variety of conditions, which influence the growth and structure of the chondro-osseous tissue leading to dysgenesis and malformation of skeletal architecture.[1],[2] One of the commonest congenital abnormalities of limbs is congenital talipes equinovarus (CTEV) of the foot also, known as clubfoot. It is characterized by hindfoot varus, forefoot adducts, cavus, and equines.[3] CTEV (clubfoot) is a multifactorial malformation, which manifests with a broad spectrum of variability in clinical features and genetic heterogeneity.[4] It may exists as an isolated entity or associated with other clinical features and is classified based on clinical presentation.[5] However, in most of the cases, CTEV presents as an isolated birth defect of the foot.[6] Rarely, it is presented in association with other congenital malformation like distal arthrogryposis, congenital myotonic dystrophy, myelomeningocele, and others.[7] The exact etiological factors involved in the development of CTEV is unknown. However, secondary CTEV with similar clinical presentation may be due to neurovascular abnormalities.[8] For the initial correction of the CTEV, the Ponseti technique of the clubfoot gives more than a 90% success rate.[9] However, regardless of the methods used for CTEV treatment, it has a strong tendency to relapse.[9]
While isolated CTEV is a more common form of congenital foot deformity. The birth prevalence is as low as 0.5 to as high as 2.0 cases/1000 live births. Environmental and genetic factors are the major contributors to high prevalence. Previous data collected from early amniocentesis showed an association of oligohydramnios as a cause of the development of CTEV.[10] Later, it was proved by analyzing the statistical data on affected CTEV individuals that, most of the environmental factors such as oligohydramnios, parental age, parental education, the season of birth, maternal anxiety or depression, use of alcohol during pregnancy have no role in the development of this anomaly.[11],[12],[13] Moreover, there is evidence signifying the role of maternal smoking during pregnancy, which increases the risk for the development of CTEV during intrauterine life.[6],[14] However, a strong family history of CTEV with maternal smoking during pregnancy increases the risk of having CTEV twenty times.[15] This observation supports the role of the involvement of the candidate genes vulnerable to a mutation in the presence of maternal smoking.
Studies based on pedigree analysis have shown that monogenic CTEV does exist, and the phenotype segregates in the families both in an autosomal dominant as well as recessive manner.[3] However, so far, only a small number of genes have been discovered in families with inherited limb defects. Nonconfirmatory variants in genes, including TBX4, PITX1, FLNB, HOXD12, HOXD13, HOXA9, have been shown, with limited evidence, as an underlying cause of the CTEV.[16] Vigorous attempts have been made so far to understand the pathobiology underlying inherited limb defects and to categorize the skeletal deformities to simplify the diagnosis.[14] However, there is a lack of consistency of phenotypic manifestation along with a bizarre clinical picture of the musculoskeletal syndrome and the precise molecular players involved in generating these musculoskeletal disorders are missing.
The available literature has revealed that CTEV might occur because of genetic defects in genes involved in limb development. However, the mutation in genes(s) underlying CTEV has yet to be identified. Pakistani population is highly consanguineous and families segregating clubfoot with autosomal recessive inheritance do exist. Therefore, gene sequencing might identify biallelic mutation as an underlying cause of clubfoot.
The present study aims to screen four genes associated with the isolated form of CTEV to detect the mutation, if any, underlying CTEV in families from Pakistan. The study will increase our knowledge of genetic variation underlying the CTEV and provide influential insight into the etiologic pathways to assess the genetic risk in the families and accurate genetic diagnosis.
Materials and Methods
Study subjects
Two inbred Pakistani families segregating CTEV were evaluated. Pedigrees were drawn and peripheral blood samples were collected from the third and fourth generation for genetic analysis [Figure - 1]. A total of four individuals were affected two in each family at the time of sample collections. The affected individuals were already diagnosed by consultant orthopedics from the Department of Orthopedics of Lady Reading Hospital Peshawar, Pakistan. These individuals were referred for the treatment to the clubfoot clinic at the Pakistan Institute of Prosthetic Sciences (PIPOS). Hence, at the time of our study, all the affected individuals were under a strict regime of serial foot casting using the Ponseti technique.
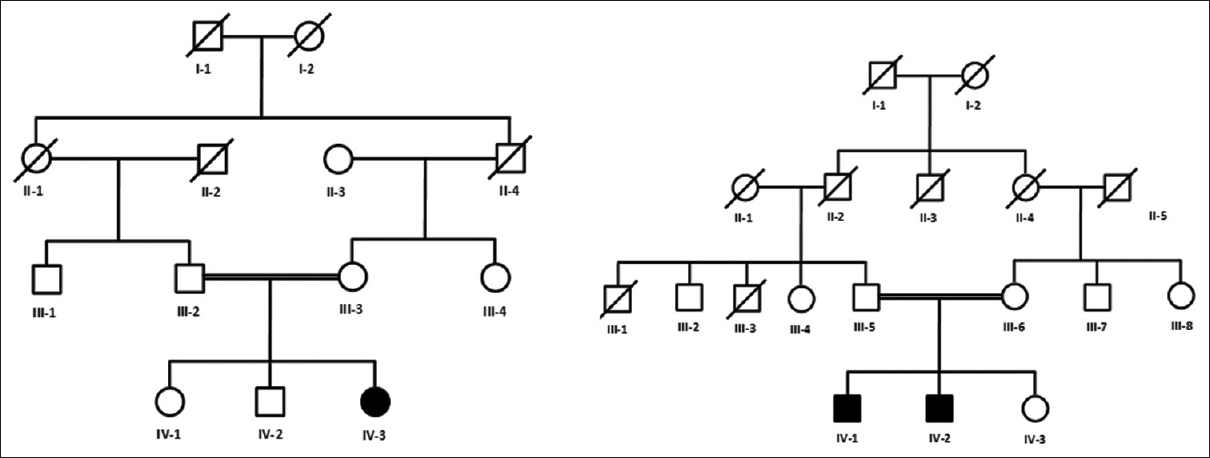 |
| Figure 1: Pedigree charts of two Pakistani kindred segregating congenital talipes equinovarus in an autosomal recessive manner. Circles represent females, squares represent males, and filled symbols represent affected individuals. Double lines between symbols indicate consanguineous marriage. Five individuals (III-2, III-3, IV-1, IV-2, and IV-3) from family A and five individuals (III-5, III-6, IV-1, IV-2, and IV-3) from family B were available for the present study |
The pedigree [Figure - 1] analysis provided conclusive evidence of the autosomal recessive inheritance of the phenotype, while affected individuals were considered homozygous for the mutant allele because of consanguineous marriages.
Extraction of genomic DNA and Sanger sequencing
Peripheral blood samples were collected from the four symptomatic and six apparently normal individuals of the two families in an ethylene diamine tetra-acetic acid-containing tubes. Total nucleic acid (DNA/RNA) was isolated using the PureLink™ Genomic DNA Mini Kit (Thermo Fisher Scientific Inc., Waltham, Massachusetts, United States). Polymerase chain reaction (PCR) was carried out in a 0.2 ml Eppendorf® PCR tubes (Sigma-Aldrich Chemie GmbH, Eschenstrasse 5, 82024 Taufkirchen, Germany) in a total volume of 50 μl. The PCR master mix (ReadyMix™ Taq PCR Reaction Mix) contained 40 ng of human genomic DNA, 25 μl of ReadyMix (Sigma-Aldrich Chemie GmbH, Eschenstrasse 5, 82024 Taufkirchen, Germany) and 20 pmol of each forward and reverse primer (Macrogen Inc., GaSan-Dong, 153-801 GeumCheon-Gu, Seoul, Korea). The thermal cycling conditions used for the amplification of genomic targets included 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, 55°–58°C for 1 min, 72°C for 1 min and a final extension at 72°C for 10 min. PCR reaction mix was amplified using Eppendorf Mastercycler nexus (USA-Scientific Inc., 346 SW 57th Ave, Ocala, FL 34474 USA). Amplified PCR products were visualized on 2% agarose gels. DNA extraction, PCR amplification and sequencing were performed in the research center of Khyber Medical University (KMU). ABI310 genetic analyzer was used for generating sequencing chromatograms.
Results
Clinical description of families
Families A and B were recruited from the Gulbahr region and Hayatabad regions of district Peshawar, respectively. The family members follow the culture of the old tribal system with the head of the family traditionally prefer their children to get married within the tribe. Hence, consanguineous marriages are commonly practiced. The pedigrees of families covering four generations show an autosomal recessive mode of inheritance of the CTEV. All individuals were clinically examined, and no history of any other congenital anomaly was observed.
In family A, two individuals (IV-2 and IV-3) were affected with CTEV, while others were normal. Affected individual (IV-2) is a 2-year-old male diagnosed as a bilateral CTEV with the lateral border of each foot was longer and convex, while the medial border of the foot was short and concave with creases. Small creases were also noted on the posterior side of the ankle. He is under strict serial casting regime according to the Ponseti technique at the PIPOS center. Medial crease and empty heel of both feet were still present. However, other signs disappeared. Affected individual (IV-3) is a 3.5-year-old female. She was diagnosed as right foot CTEV and underwent a similar regime. Signs of CTEV were about to disappear completely. However, as relapse chances are high, she was still on regular follow-up every 3 months in the clubfoot clinic at the PIPOS center. Blood samples were collected from the two affected (IV-2 and IV-3) and three unaffected members (III-2, III-3, and IV-1) of family A.
In family B, two male individuals (IV-I and IV-2) were affected with CTEV. Affected individual (IV-1) is 2.5 years diagnosed as a bilateral CTEV. He was under strict serial casting according to the Ponseti technique at the PIPOS center. He also underwent percutaneous Achilles tenotomy as the feet were resistant to conservative management. The second affected individual (IV-2) is a 9-month-old male. He was diagnosed as right foot CTEV and underwent a similar regimen. Still, signs of CTEV such as metatarsal adducts, cavus, and equinus on both feet were apparent. Blood samples were collected from the two affected (IV-1 and IV-2) and three unaffected members (III-3, III-6, IV-3) of family B.
Sequence analysis of four known congenital talipes equinovarus genes
A total of 10 DNA samples from two families (A and B) were screened for four known genes associated with the isolated form of CTEV. A total of twenty primers flanking exons and exons-introns junctions were used for PCR amplification, followed by Sanger sequencing of hereditary CTEV related genes via dideoxy chain termination chemistry to identify variant(s) of interest underlying CTEV in both families. Three exons and exon-intron junctions of the PITX1 gene, eleven exons and exon-intron junctions of the TBX4 gene, four exons of HOXD12 gene, and two exons of the HOXA9 gene were sequenced in the affected individuals of family A (IV-2) and family B (IV-1).
A homozygous sequence variant (c.1227C>T) in exon 8 of the TBX4 gene was detected in the affected individual (IV-1) of family B. This variant was sequenced in all other available individuals of the family B as well. It is perfectly segregating with the disease phenotype in family B. The variant is present in a homozygous state in both affected individuals and in a heterozygous state in both parents. The unaffected individuals showed wild type sequence. The variant (c.1227C>T) changes the codon GAC to GAT at position 408 (p. Asp408Asp). However, this variant did not lead to any amino acid change in the protein sequence as GAC and GAT code for the same amino acid (aspartate). Moreover, the genome AD database search revealed that this synonymous variant is present in high frequency in the general population. No other sequence variants were identified, suggesting that the mutation may be present in the regulatory region of the gene.
Discussion
Genetic studies in congenital talipes equinovarus
In the pediatric age group, the most common, challenging, and complicated disorder of the musculoskeletal system is the development of CTEV. The genetic component involved in the development of the CTEV is predominant. While depending on its severity, neuromuscular or vascular system hypoplasia may predominate later in the development of the CTEV.[17] CTEV occurs as an isolated (nonsyndromic) birth defect in 80% of the cases without having other associated malformation.[8]
Around 24%–50% of the CTEV cases are reported with a positive family history.[18] As the genetic factors have a predominant contribution in the etiology of this deformity, which is the cause of treatment failure and reoccurrence in the families affected with CTEV.[19] Pedigree analysis of multiple affected individuals suggesting the key role of a single gene, with both autosomal recessive and dominant patterns of inheritance.[20] The involvement of genetic contribution to the development of CTEV is different from individual to individual and is expected to be small to moderate in size and vary in families and different populations.[21]
In this study, we focused on the clinical and genetic characterization of two consanguineous families (A and B) showing the hallmark phenotype of CTEV. Clinical examination of affected individuals from both families confirmed the diagnosis of CTEV. DNA extraction from all available members followed by primer designing for four CTEV associated genes (PITX1, TBX4, HOXA9, and HOXA12) and Sanger sequencing revealed no variant in the DNA samples of the members of family A. However, a homozygous sequence variant (c.1227C>T) in the exon eight of the TBX4 gene was identified in family B. The variant is synonymous and therefore did not cause any amino acid change in the protein sequence.
Therefore, no disease-causing sequence variant was identified in all four genes sequenced. The gene PITX1-TBX4 genes are responsible for the transcriptional pathway of initial limb development. Previous studies on PITX-TBX4 genes support the involvement of these genes in the development of CTEV.[21],[22] In a multigenerational family segregating CTEV in an autosomal dominant manner Gurnett et al., 2008[23] identified a missense mutation (p. E130K) in the PITX1 gene that was located in a highly conserved domain. The clubfoot-like phenotype was also observed in the mice with Pitx 1 haploinsufficiency.[5],[23] This shows the importance of PITX1-TBX4 in the developmental pathway of CTEV.[24] Role of PITX1 and TBX4 in the hind limb is not known. Both PITX and TBX4 are mainly involved in the development of the forelimb in model animals. This strengthen the hypothesis that these genes play a major role in the development of foot in human.[25] This study and others have shown that multiple families with isolated CTEV failed to show mutations in skeletal muscles contractile genes and other CTEV associated genes, including Filamin B (FLNB) gene.[17],[26]
Muscle development is a complex process that involves spatiotemporal expression of multiple genes during intrauterine life. Molecular alteration of this process may lead to muscle anomalies such as hypoplasia and atrophy of muscle, which are the characteristic features of CTEV.[27] During the early stages of myogenesis, these changes are highly controlled with their exactly related specific genes, which play a vital role in the development of limb and foot.[28] Studies conducted based on candidate gene association analysis suggest that apoptosis-related genes, homeobox A and D (HOXA and HOXD) genes, and genes involved in muscle contraction are major contributors in the development of CTEV.[29],[30],[31] During embryonic life, the homeobox transcription factor gene, HOXA9, plays a role in coordinated patterning and differentiation of muscle, tendon, and cartilage of the hindlimbs.[32],[33]HOXA9 is also known to control the expression of genes by regulating the transcription factor such as LBXI. LBXI plays an important role in the migration of muscle precursor cells during the development of hind limb.[34] A decreased promotor activity with an alternate allele of HOXA9 ( rs3801776) was found due to a single nucleotide polymorphism (SNP).[35] This may lead to a change in the structure of muscle, calf size, and the disordered functional composition of muscle. All these components were reported in the development of CTEV. Moreover, genotypes of 4 SNPs in 84 CTEV patients were analyzed, and it was found that rs847154 located in 5' flanking sequence of HOXD12 gene is associated with CTEV.[36] We sequenced the two exons of the HOXA9 and four exons of the HOXD12 gene in affected individuals of family A (IV-2) and family B (IV: 1). However, no sequence variant was identified.
We did not identify any pathogenic sequence variant(s) in four CTEV candidate genes (PITX, TBX4, HOXA9, and HOXD12). This suggests that the mutation may be present in the regulatory region of these genes. Whole-exome sequencing (WES) might reveal the pathogenic variant underlying CTEV in these patients.
Recently, next-generation sequencing (NGS), including whole-genome sequencing and WES, is becoming more common in clinical diagnosis as well as gene identification in genetic diseases. WES is cost-efficient, and simple bioinformatics approaches have the potential to identify the underlying genetic variants in inherited diseases. WES is becoming a gold standard diagnostic tool for the discovery of the culprit gene while studying the undiagnosed genetic disease.[37] This particularly efficient method for the identification of undiagnosed disease-associated genes can be more useful when applied in combination with genetic linkage analysis.[35] Future studies, including whole-genome SNP genotyping and WES on these samples may lead to interesting results in family A and B.
Conclusion
Based on our data, it is highly unlikely that mutations in the coding part of these genes are the cause of CTEV in the Pakistani population. However, a study with a large sample size will justify this conclusion. Further research using the NGS approach followed by segregation analysis is necessary for the detection of potential sequence variants underlying CTEV. It is important to search for the culprit gene involved in the CTEV because it is difficult to treat the disease properly until the true etiology and pathological mechanism is known. Identification and characterization of novel gene discovery involved in CTEV will provide accurate genetic counseling for families at risk as well. This will also provide the advancement in the management plane, especially in treatment-resistant cases, as not all the CTEV cases respond well to the available treatment options. The personalized case-based approach between the genetic abnormality and CTEV malformation will be a huge advancement in the available management plan.
Ethical consideration
This study was approved by the Graduate Study Committee and the Advanced Study and Research Board of KMU Peshawar, Pakistan.
Acknowledgment
We are thankful to the members of the two families for giving us permission to conduct the genetic studies and publish their clinical and genetic data in this manuscript.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form the patients have given their consent for their images and other clinical information to be reported in the journal. The patients understand that their names and initials will not be published and due efforts will be made to conceal their identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Author's contribution
YNK and ZH recruited families and performed the clinical evaluation the DNA extraction. YNK and UK performed PCR and Sanger sequencing. YNK wrote the initial draft. SB performed data analysis and revised the manuscript. All authors have seen and agreed to the content of the manuscript. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
| 1. | Duboc V, Logan MP. Regulation of limb bud initiation and limb-type morphology. Dev Dyn 2011;240:1017-27. [Google Scholar] |
| 2. | Panda A, Gamanagatti S, Jana M, Gupta AK. Skeletal dysplasias: A radiographic approach and review of common non-lethal skeletal dysplasias. World J Radiol 2014;6:808-25. [Google Scholar] |
| 3. | Basit S, Khoshhal KI. Genetics of clubfoot; recent progress and future perspectives. Eur J Med Genet 2018;61:107-13. [Google Scholar] |
| 4. | Ponseti IV, Campos J. The classic: Observations on pathogenesis and treatment of congenital clubfoot. 1972. Clin Orthop Relat Res 2009;467:1124-32. [Google Scholar] |
| 5. | Sadler B, Gurnett CA, Dobbs MB. The genetics of isolated and syndromic clubfoot. J Child Orthop 2019;13:238-44. [Google Scholar] |
| 6. | Pavone V, Chisari E, Vescio A, Lucenti L, Sessa G, Testa G. The etiology of idiopathic congenital talipes equinovarus: A systematic review. J Orthop Surg Res 2018;13:206. [Google Scholar] |
| 7. | Lochmiller C, Johnston D, Scott A, Risman M, Hecht JT. Genetic epidemiology study of idiopathic talipes equinovarus. Am J Med Genet 1998;79:90-6. [Google Scholar] |
| 8. | Herceg MB, Weiner DS, Agamanolis DP, Hawk D. Histologic and histochemical analysis of muscle specimens in idiopathic talipes equinovarus. J Pediatr Orthop 2006;26:91-3. [Google Scholar] |
| 9. | Rasheed N, Khani GM, Zaidi IH. Club foot after treatment; pattern and causes of relapses with Ponseti technique. Prof Med J 2018;25:514-9. [Google Scholar] |
| 10. | Ponseti IV, Zhivkov M, Davis N, Sinclair M, Dobbs MB, Morcuende JA. Treatment of the complex idiopathic clubfoot. Clin Orthop Relat Res 2006;451:171-6. [Google Scholar] |
| 11. | Dodwell E, Risoe P, Wright J. Factors associated with increased risk of clubfoot: A Norwegian national cohort analysis. J Pediatr Orthop 2015;35:e104-9. [Google Scholar] |
| 12. | Gizzo S, Noventa M, Vitagliano A, Dall'Asta A, D'Antona D, Aldrich CJ, et al. An update on maternal hydration strategies for amniotic fluid improvement in isolated oligohydramnios and normohydramnios: Evidence from a systematic review of literature and meta-analysis. PLoS One 2015;10:e0144334. [Google Scholar] |
| 13. | Werler MM, Yazdy MM, Mitchell AA, Meyer RE, Druschel CM, Anderka M, et al. Descriptive epidemiology of idiopathic clubfoot. Am J Med Genet A 2013;161A: 1569-78. [Google Scholar] |
| 14. | Ippolito E, De Maio F, Mancini F, Bellini D, Orefice A. Leg muscle atrophy in idiopathic congenital clubfoot: Is it primitive or acquired? J Child Orthop 2009;3:171-8. [Google Scholar] |
| 15. | Honein MA, Paulozzi LJ, Moore CA. Family history, maternal smoking, and clubfoot: An indication of a gene-environment interaction. Am J Epidemiol 2000;152:658-65. [Google Scholar] |
| 16. | Barrow JR, Thomas KR, Boussadia-Zahui O, Moore R, Kemler R, Capecchi MR, et al. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev 2003;17:394-409. [Google Scholar] |
| 17. | Gurnett CA, Alaee F, Desruisseau D, Boehm S, Dobbs MB. Skeletal muscle contractile gene (TNNT3, MYH3, TPM2) mutations not found in vertical talus or clubfoot. Clin Orthop Relat Res 2009;467:1195-200. [Google Scholar] |
| 18. | Miedzybrodzka Z. Congenital talipes equinovarus (clubfoot): A disorder of the foot but not the hand. J Anat 2003;202:37-42. [Google Scholar] |
| 19. | Chapman C, Stott NS, Port RV, Nicol RO. Genetics of club foot in Maori and Pacific people. J Med Genet 2000;37:680-3. [Google Scholar] |
| 20. | Sreenivas T, Nataraj AR. Parental consanguinity and associated factors in congenital talipes equinovarus. Foot (Edinb) 2012;22:2-5. [Google Scholar] |
| 21. | Alvarado DM, Aferol H, McCall K, Huang JB, Techy M, Buchan J, et al. Familial isolated clubfoot is associated with recurrent chromosome 17q23.1q23.2 microduplications containing TBX4. Am J Hum Genet 2010;87:154-60. [Google Scholar] |
| 22. | Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Pitx 1 and Pitx 2 are required for development of hindlimb buds. Development 2003;130:45-55. [Google Scholar] |
| 23. | Gurnett CA, Alaee F, Kruse LM, Desruisseau DM, Hecht JT, Wise CA, et al. Asymmetric lower-limb malformations in individuals with homeobox PITX1 gene mutation. Am J Hum Genet 2008;83:616-22. [Google Scholar] |
| 24. | Peterson JF, Ghaloul-Gonzalez L, Madan-Khetarpal S, Hartman J, Surti U, Rajkovic A, et al. Familial microduplication of 17q23.1–q23.2 involving TBX4 is associated with congenital clubfoot and reduced penetrance in females. Am J Med Genet A 2014;164A: 364-9. [Google Scholar] |
| 25. | Logan M, Tabin CJ. Role of Pitx 1 upstream of Tbx4 in specification of hindlimb identity. Science 1999;283:1736-9. [Google Scholar] |
| 26. | Yang H, Zheng Z, Cai H, Li H, Ye X, Zhang X, et al. Three novel missense mutations in the filamin B gene are associated with isolated congenital talipes equinovarus. Hum Genet 2016;135:1181-9. [Google Scholar] |
| 27. | de Andrade M, Barnholtz JS, Amos CI, Lochmiller C, Scott A, Risman M, et al. Segregation analysis of idiopathic talipes equinovarus in a Texan population. Am J Med Genet 1998;79:97-102. [Google Scholar] |
| 28. | Alvarado DM, Buchan JG, Frick SL, Herzenberg JE, Dobbs MB, Gurnett CA. Copy number analysis of 413 isolated talipes equinovarus patients suggests role for transcriptional regulators of early limb development. Eur J Hum Genet 2013;21:373-80. [Google Scholar] |
| 29. | Weymouth KS, Blanton SH, Powell T, Patel CV, Savill SA, Hecht JT. Functional assessment of clubfoot associated HOXA9, TPM1, and TPM2 variants suggests a potential gene regulation mechanism. Clin Orthop Relat Res 2016;474:1726-35. [Google Scholar] |
| 30. | Wang Y. Relationship between HOX gene and pediatric congenital clubfoot. Exp Ther Med 2018;15:4861-5. [Google Scholar] |
| 31. | Shrimpton AE, Levinsohn EM, Yozawitz JM, Packard DS Jr., Cady RB, Middleton FA, et al. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot-Marie-Tooth disease. Am J Hum Genet 2004;75:92-6. [Google Scholar] |
| 32. | Raines AM, Magella B, Adam M, Potter SS. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev Biol 2015;15:28. [Google Scholar] |
| 33. | Alvarado DM, McCall K, Hecht JT, Dobbs MB, Gurnett CA. Deletions of 5' HOXC genes are associated with lower extremity malformations, including clubfoot and vertical talus. J Med Genet 2016;53:250-5. [Google Scholar] |
| 34. | Dobbs MB, Gurnett CA. Update on clubfoot: Etiology and treatment. Clin Orthop Relat Res 2009;467:1146-53. [Google Scholar] |
| 35. | Alharby E, Albalawi AM, Nasir A, Alhijji SA, Mahmood A, Ramzan K, et al. A homozygous potentially pathogenic variant in the PAXBP1 gene in a large family with global developmental delay and myopathic hypotonia. Clin Genet 2017;92:579-86. [Google Scholar] |
| 36. | Wang LL, Jin CL, Liu LY, Zhang X, Ji SJ, Sun KL. Analysis of association between 5' HOXD gene and idiopathic congenital talipes equinovarus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2005;22:653-6. [Google Scholar] |
| 37. | Belkadi A, Bolze A, Itan Y, Cobat A, Vincent QB, Antipenko A, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A 2015;112:5473-8. [Google Scholar] |
Fulltext Views
3,295
PDF downloads
1,414