Translate this page into:
Precision oncology innovation: Sarcoma as an example
Corresponding Author:
Abdul-Mohsen G Alhejaily
Department of Basic Medical Science, Faculty of Medicine, King Fahad Medical City, Riyadh
Saudi Arabia
aalhejaily@kfmc.med.sa
| How to cite this article: Alhejaily AMG, Salem RO. Precision oncology innovation: Sarcoma as an example. J Musculoskelet Surg Res 2020;4:3-8 |
Abstract
Patients suffering from cancer will be extensively categorized for gene mutations and other genetic aberrations, and their treatment will uniquely be based on molecular profiling instead of their cancer being initiated or detected in the body. This approach is part of what is called precision medicine (PM). The better molecular understanding of cancer that we are gaining and applying toward the development of new therapies will be applicable to cancer prevention and screening. The field of PM in oncology or precision oncology (PO) is an appropriate choice to begin the initiative of PM. The integration of genomic information from tumors with clinical information, including treatment response and patient outcomes, will create a helpful resource to enhance anticancer therapeutics that are more effective and less toxic than the traditional treatments. Sarcomas are a heterogeneous group of tumors that display a remarkable heterogeneity, with more than 50 subtypes identified so far. Enhancements in the high throughput genomic sequencing have contributed to the discovery of genomic events associated with these heterogeneous tumors and the underlying biology, have opened up paths to discover molecular targeted therapeutics, and improve the clinical decision-making processes. This review will focus on how the integration of the PO approach will affect the drug development in sarcomas and strategies for optimizing finding of novel diagnostic, prognostic, and predictive biomarkers in addition to developing more-effective personalized therapeutic approaches for patients with sarcoma.Introduction
It is well known now that cancer is a genetic disease; in other words, it is caused by changes in the DNA that regulate how cells grow and divide. These changes can be inherited, but most appear during a person's life, mostly due to exposure to carcinogens that damage DNA.[1] We live in the era of genomic medicine and the integration of next-generation sequencing (NGS) into clinical oncology.[2] The combination of the advanced NGS technology and computational data analysis methods has revolutionized our understanding of the genomic underpinnings of cancer development and progression at the molecular level. The past decade brought significant achievements in the field of cancer research, driven by rapidly evolving technologies and reduction in the costs of NGS technologies. Scientists who specialized in cancer genomics and molecular biology, together with bioinformatics, are the core of the research unit, and their cooperation as a team is vitally important to develop and translate the basic research findings from the laboratories into clinical practice.[3] This multidisciplinary collaboration in cancer research and care will lead to novel findings of more appropriate treatment approaches as well as the development of individual treatment plans for each patient based on his/her unique genomic variations.[4] Cancer genetic alterations can help choose a more precise personalized treatment plan which is known as “precision oncology” (PO).
As our understanding of the tumor biology plays a crucial role in understanding of how cancer initiation, transformation, and progression grows continuously, new opportunities for the early detection and intervention will certainly be available.[5] In the near future, oncology research will need to have a much deeper understanding of precise “tumor biology” on a molecular level for each cancer patient using comprehensive genomic profiling, which is now become increasingly important, especially with targeted cancer therapeutics.[6] Investment in new technologies in cancer research will eventually provide significant scientifically novel approaches in cancer treatment and prevention.[7] Moreover, advancement in clinical research and clinical trials offers a quality of cancer care, provides patients with the opportunity to take a vigorous role in their treatment plans, and allows improving cancer treatment for future patients. Cancer research is becoming more focused on patient-oriented care.[8]
At the level of the tumor biology, cancer is described as a “genomic disease.” This is because mostly the tumor cells have encountered irreversible changes “mutations” in one or more of the “key genes” that control the cell cycle.[9],[10] These mutations will lead to abnormal cellular changes, such as an increase or decrease in their activity and an alteration in the copy number of certain vital genes, which often either increases their activity or upregulating oncogenes or decreases their activity or downregulation of tumor suppressor genes.[11] Furthermore, the genetic changes in key genes are critical to the initiation of the carcinogenesis process, and such cells are valuable as tumor markers in certain cancers.[12] Molecular alterations involved in carcinogenesis are very diverse, as are the mechanisms by which cellular functions may be altered. Mutations in genes controlling the DNA synthesis process will change the genetic stability of cells and then contribute to the progression of neoplasia, which, in turn, leads to the accumulation of genetic alterations in evolving clones. Therefore, gene mutations that stimulate a selective growth advantage appear to play a key role in carcinogenesis.[13]
Advancement of our knowledge of cancer genomics has expanded our understanding of the genetic basis of cancer and the identification of genomic markers of cancer risk in population studies to characterize the biological mechanisms of cancer at the genetic level in the laboratory.[14] Scientists have been studying cancer genetics in efforts to elucidate the genetic subtypes of various cancer and translating that knowledge into new ways for creating a novel therapeutic approach, which paves the way toward future precision medicine (PM) strategies.[15]
Precision Medicine
In addition to what was mentioned in the introductory section, PM initiatives use genomic information from the individual patient to provide a customized health care.[16] These might include but not limited to the disease's diagnosis, treatment, and prognosis. Exclusively, in cancers, data obtained from the NGS system are utilized in personalized diagnosis and prognosis of diseases, defining targeted therapy, and evaluating the appropriateness of a patient to be part of a clinical trial.[17] For example, now NGS lets us look at any number of genes that are engaged in the oncogenic process of a patient's tumor. The usage of NGS platforms is more efficient and provides a much better resolution compared to microarrays.[18] Hence, its adoption is spreading fast on a global basis.
The process of variant categorization and reporting often needs settling multiple lines of evidence for evaluating the system. Moreover, widespread implementation of NGS technologies has generated a massive number of variants to be used in the analysis and categorization many of diseases based on their molecular heterogeneity.[19] There are numerous data sources, the so-called “Big Data” which contains a wealth of information about the clinical relevance of genomic variants.[20] In addition, there is a wide array of algorithms that help to assess the presence of cancer initiation and progression. Molecular cancer diagnostics is a quickly evolving discipline. New papers describing treatment options and new associations between genes and cancers are published daily.[21] In addition, new clinical trials are opened regularly. At present, oncologists are under the obligation to apply state-of-the-art knowledge in their diagnostic and therapeutic decision making. On the other hand, we have reached a level of complexity in the available data, information, and knowledge where the manual development of a defendable clinical report is extremely difficult.[21] Soon, software-aided decision-making will be a feasible option to deal with this complex matter. PM trials require sophisticated and expensive technologies and clinical processes that have not typically been a part of most clinical trials recently.
Molecular Profiling of Tumors
Using state-of-the-art technologies and unrivaled expertise in the molecular characterizations of tumors, tumor profiling (TP) supports researchers to accelerate progress toward precision therapies.[21] TP provides access to a range of cutting-edge techniques for genomic analysis such as NGS platforms and cancer biomarkers, and much of the knowledge about this set of diseases has come from studies in experimental models in the laboratory.[22] Over the past couple of decades, novel genetic methods have been used to bread mice that develop cancer in a predictable way in a variety of tissues under the influence of mutant genes to mimic certain cancers found in humans.[22]
Technological molecular profiling advances are making it easier to study the consequences of specific gene mutations and potential therapeutic approaches. In addition, methods for growing cancer cells, classifying cancers according to patients' genomic profiles, have helped in the development of effective cancer treatment, which can be directed against the specific genetic aberrations in each patient's tumor [Figure - 1].[23] Such classification is important both for cancer treatment diagnosis and for research that can lead to new improved targeted treatments that may lead to a better outcome for patients with cancer in future.
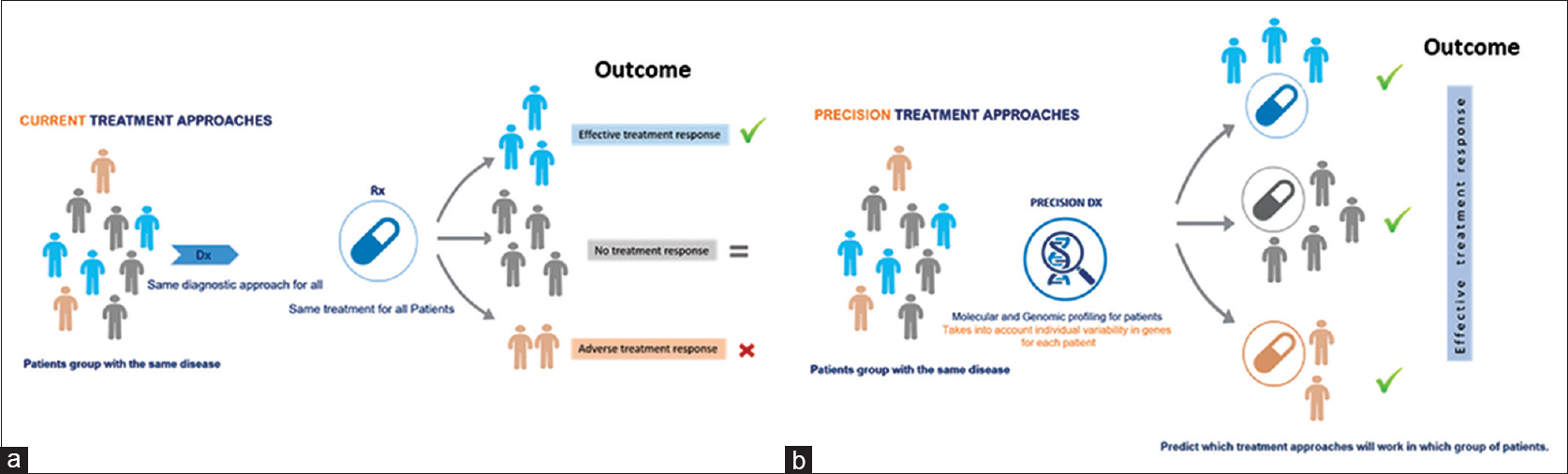 |
| Figure 1: An illustration of the Precision medicine a paradigm in treatment delivery regimens. Precision medicine approaches are enabled by genomic data to influence from direct and indirect sources to provide a more general view of an individual patient based on his/her genomic-makeup and then applying precision medicine into mainstream clinical decision will eventually brining much better targeted therapies. (a and b) compare the current approach in which all patients with same disease treated equally, where in precision medicine approach the treatment take into account the disease heterogeneity for each patient |
Precision Oncology
The fundamental role in the delivery of PO research has evolved diagnostic tools and tests that dealt with the occurrence of specific genomic variations (biomarkers) in the patient's tumors. These biomarkers can anticipate the response to specific targeting agents (predictive biomarkers) or provide prognostic information about the disease outcome (prognostic biomarkers).[24] These different types of biomarkers are helping in matching patients with the available therapeutic agents (targeted therapy) or therapeutic regimens (tailored treatment “chemotherapy”).[25] There are a number of technologies that are presently used for diagnostic assessment and genomic profiling for cancer patients such as tumor biomarkers, immunohistochemistry (IHC), cytogenetics, array comparative genomic hybridizations, and most importantly NGS platforms. Understanding the molecular changes in cancer will hopefully encourage the development of targeted drugs (tailored treatment) that will precisely target key proteins implicated in the initiation and progression of cancer. This therapeutic approach is often referred to as PO.[26]
Another aspect of PO is the development of specific molecular assessments or biomarkers analysis often called companion diagnostics (CDx), which means a specific diagnostic test for each therapeutic product. The CDx can predict patients who are most likely to benefit from a particular targeted therapy,[27] for example, the FDA conditional approval of “vemurafenib” (Zelboraf) for patients with advanced melanoma whose tumors “harbor” a frequent mutation in the sarcoma viral oncogene homolog B1 protooncogene (BRAF) oncogene only, for patients to perform an FDA-approved CDx test prior to taking this drug. Similarly, to this far, erlotinib (Tarceva), which targets the epidermal growth factor receptor (EGFR) gene mutation, in lung adenocarcinoma and trastuzumab (Herceptin), which targets human epidermal growth factor receptor 2 (HER2) mutation in lung adenocarcinoma and breast cancer. This information contributes to the predictability that certain pharmaceuticals or therapeutic approaches will provide long-term solutions to disease in an individual with cancer.[28]
Even though a significant number of genomic alterations that lead to the initiation, progression, and transformation across different cancer types have been detected and validated so far, some tumors have not been fully elucidated yet.[29] In addition, studies compare the genomic variations from tumor and nontumor cells from the same patient using technology such as laser capture microdissection which allow researchers to obtain subpopulations of tissue cells under direct microscopic visualization for several downstream analysis. As an example of these analysis, DNA-genotyping and signaling pathway analysis from highly heterogenous tumor tissue.[30] Furthermore, the analysis of DNA, RNA, and protein derived from heterogeneous tissue samples has transformed pathology and resulted in the determination of diagnostic and prognostic biomarkers that will have a great influence on clinical practice.[31]
The abundance of data gained from the cancer genomics gradually will be integrated with patients' medical records and clinical data. This integration approach between clinical and genomic data is already used now to develop tailored therapeutic approaches, enhance the methods of predicting prognosis, and response to treatment. In addition, the integration helps identify the molecular subtype of the tumor, which has different molecular signatures and prognostic information.
A few challenges normally rise in comprehensive cancer genomic analysis of cancer. There are many genomic alterations that drive cancers, and therefore, this will be a challenge in this field.[31] Another challenge is acquiring high-quality biological samples needed for genomic studies, especially for tumor types that are usually rare and of low prevalence. Samples such as formalin-fixed paraffin embedded, or where the DNA/RNA are highly degraded, constitute a challenge that should be considered when using this type of samples.[32] Another challenge is managing and analyzing the vast amounts of data (big data), which is generated from the genomic analysis.[33] Moreover, further advancement of microarray and large-scale high-throughput NGS sequencing to map the landscape of the cancer genome will frequently discover new deleterious changes linked to specific type/subtype of cancer and generate big data that need to be translated to the clinical domain. Hence, integrating the results from several analyses helps scientists to gain a better understanding of cancer, but it needs good infrastructure, computational resources, and tools to store, process, and analyze the data. The accessibility of these data, and the insights they may offer into the underline biology of the tumor, has many advantages toward the PO, prevention, diagnosis, and treatment based on the molecular signature of each patient.
Ultimately, the PO must be merged with modern, effective treatment strategies by robust clinical testing and verification. Furthermore, clinical trials must utilize the ingrained boundaries of PO in targeting molecular alterations within various tumor entities and should be based on their evidence-based clinical utility. Taking all these into consideration will bring the era of PO into the mainstream of health care.
Precision Oncology for Sarcoma
Sarcomas are a heterogeneous group of rare malignancies that carry a remarkable heterogeneity clinically and histopathologically, with more than 50 subtypes recognized so far.[34] Moreover, sarcomas are malignant mesenchymal that characterized by a distinguished high molecular heterogeneity, which reflects the disease biological complexity and leads to substantial challenges in their diagnosis and clinical management. At present, the diagnosis of sarcoma is based on morphology through IHC and clinicopathological correlation. However, the advances in NGS technology have resulted in the more precise discovery of actionable genetic events in these groups of tumors, which in addition enhance our understanding of the underline biology and the complexity of this disease and have opened up opportunities for more beneficial and effective molecularly targeted therapy.[34] Cytogenetically, a binary discrepancy among sarcomas with a simple chromosomal karyotype versus those with a complex karyotype has been offered a straightforward context of some value of cytological significance of chromosomal instabilities to detect and diagnose the different subtypes of this disease.[34] Besides, the molecular association of these cytogenetic alterations is frequent and genomic rearrangements and actionable gene mutations for sarcoma subtypes with different cytogenetic features. In addition, various genomic events have been identified including oncogene amplifications and gene fusions, for those with a complex karyotype. Biologically, oncogenic mechanisms in sarcomas are easily recognized with simple karyotype rather than the complex ones and those fall typically into one broad category: sarcomas with transcriptional deregulation in signaling pathways that related to sarcomas initiation. However, in sarcomas with complicated karyotypes, which usually do not harbor single-driver genetic alterations and not display specific molecular changes that promote the oncogenesis, such as genomic instability mostly involved in cell cycle deregulation and promotion of uncontrolled cellular growth. These types of sarcomas are difficult to detect by simple cytogenetic tests and required more sophisticated techniques.[34],[35]
In 2017, National Cancer Institute (NCI)-funded researchers at the University of Texas Health Science Center at San Antonio reported findings from a study of the fusion oncoprotein EWS-FLI1, which drives approximately 85% of Ewing sarcomas.[35] These tumors occur mainly in children and young adults and are found most often in the bones. The researchers discovered that EWS-FLI1 increases tumor cell production of an enzyme called pappalysin-1 (PAPPA), which breaks down certain proteins called insulin-like growth factor-binding proteins (IGFBPs). The breakdown of IGFBPs releases the hormone/insulin-like growth factor into the local environment where it promotes cancer cell growth.[36]
The researchers also showed that inactivating PAPPA might be an effective strategy in treating Ewing sarcoma.[37] This will increase the focus on the basic medical research related to this mediator, and other “fusion oncoproteins” might optimistically lead to new therapeutic approaches of these cancer types. Some investigators are moving beyond searching for driver mutations and instead are asking whether NGS can predict response or resistance to therapy; others are creating expression profiles that go beyond individual genes. NGS has the potential to become, in the coming years, an established platform of choice for researchers who are studying sarcomas.[38],[39]
To date, many investigators have attempted to identify recurrent aberrations using NGS. A large variety of identified mutations in cancer-associated pathways are predominantly secondary mutations that occur later in the course of tumorigenesis. They may be responsible for accelerated growth in advanced disease but are unlikely to be the sole cause of the malignancy. Identification of these mutations is important, and with so much diversity in involved pathways, an individualized approach is necessary for proper sarcoma treatment.
Opportunities for Greater Future Progress
Now, there is a huge effort around the world among researchers and clinicians to improve the effectiveness of the health-care services and to develop more effective treatments by collecting data and developing new tools to design and tailor health care to patients based on patient's unique genomic makeup, environment, and lifestyle. At the present time, most of the focuses are on research programs to support the unremitting growth in the areas of personalized medicine, and pharmaceutical companies around the world lead most of these research programs because of their belief in the feasibility of investing in this area. Moreover, scientists around the world are working on assembling data to uncover new findings that would improve the ability to treat patients as specifically as possible.
Despite the massive advances in these areas, there is still much more that needs to be learned, and there are a variety of PM breakthroughs that should make their way to patients within the next 5–10 years. Such thrilling progression is what has been done and need to be done for making an outstanding improvement in our perception of the mechanisms of cancer by improving our inclusive thoughtful of the molecular alterations that drive the initiation and progression of malignant tumors has been an enviable aim in cancer research over the past two decades. The most important movement in this direction was accomplished by the Cancer Genome Atlas (TCGA), which is a very ambitious program funded by the NCI and the National Human Genome Research Institute. The TCGA project revealed the primary molecular characteristics of 33 of the very common types of human cancers using an in-depth analysis of tumor samples and clinicopathologic features from 11,000 cancer patients. The outcomes from the TCGA project have guided to an improvement of our understanding of the molecular basis of cancer development and provided genome-wide information uncovering new key regulators of signaling pathways in different types of cancers.
Currently, the importance of PM as a vital part of the understanding of molecular heterogeneity of cancers needs the implementation and development of new paradigms and a reliable basis for PM integrating genomics data into clinical practice. Big data analysis of genomics data is an integral part of PM. It is intended to translate the data generated at cellular and molecular levels into clinically relevant information. Therefore, in the era of PM, big data analysis plays a crucial role in precisely examine diseases and incorporate these data with genomic variation and clinical information. Now, several automated data mining tools are being developed to extract genomic information, variations, and their association with diseases. However, translational and clinical research is still required for the development of safe and effective therapeutic approaches to improve cancer patients' outcomes and quality of life. This is pretty much the major components in future attempts to overcome.
Concluding Thoughts
Technological improvement in PM and PO will continue since the cost of whole-genome sequencing, and whole-exome sequencing has dropped from tens of millions of dollars to <$1,000 per genome today. Correspondingly, PM interventions are likely to proliferate over the next few years and will transform the way health-care services are delivered. However, health-care systems around the world will need to consider adjusting their evaluation approaches to adapt these changes in such a way that they can robustly assess the new treatment approaches and sustain the expected growing role of precision and personalized medicine for cancer treatment.
Professionally conducting clinical trials on multigene tumor signatures and develop more evidence-based prognostic and predictive molecular assays is a necessity. In addition, the development of a clinical decision approach based on multigene panel testing will pave the way for therapeutic personalized decision and the development of novel therapeutic interventions. In addition to more precise molecular subtyping of different tumors, a further generation of high-quality genomics data and integrating them with histopathological and clinical findings will open new avenues for customized therapy to match each individual patient's needs. There is an urgent need to validate some promising therapeutic approaches to PO. This endeavor depends on continued collaboration between clinicians, scientists, and patients.
Collectively, notwithstanding promising successful stories of PO as mentioned in this article, there are significant pitfalls before PO becomes a standard of care. One of the fundamental pitfalls is the translation of the promise of PM into real-world clinical practice. Precision promise will constantly and rapidly bring new treatment options in the next few years to help patients and to transform the future of healthcare. To accomplish a profound understanding of cancers and explore new, clinically reliable tools for molecular profiling for patients, we will need to analyze many more cancer genomes. Equally important, is the need to accelerate the implementation of new therapeutic tactics, we need the continuation of clinical research on novel drug designs performed on patients and more reliable models for preclinical testing, and after all, this only represents the tip of the iceberg when it comes to making PM a reality.
Ethical consideration
This is a review article that does not involve patients or patients' information.
Acknowledgment
We thank the reviewers for their careful reading and their insightful comments and suggestions.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Authors' contributions
AA conceived and designed the article's outlines and wrote the initial and final draft. RS collected, organized data, and revised the manuscript. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
| 1. | American Cancer Society. Cancer facts and figures 2017. Genes Dev 2017. [Google Scholar] |
| 2. | Daber R, Sukhadia S, Morrissette JJ. Understanding the limitations of next generation sequencing informatics, an approach to clinical pipeline validation using artificial data sets. Cancer Genet 2013;206:441-8. [Google Scholar] |
| 3. | Zhang J, Chiodini R, Badr A, Zhang G. The impact of next-generation sequencing on genomics. J Genet Genomics 2011;38:95-109. [Google Scholar] |
| 4. | Soukup T, Lamb BW, Arora S, Darzi A, Sevdalis N, Green JS. Successful strategies in implementing a multidisciplinary team working in the care of patients with cancer: An overview and synthesis of the available literature. J Multidiscip Healthc 2018;11:49-61. [Google Scholar] |
| 5. | Phay JE, Moley JF. Genetics of cancer. In: Essential Practice of Surgery: Basic Science and Clinical Evidence. 2nd ed. Springer: New York; 2008. p. 1901-24. [Google Scholar] |
| 6. | Imran A, Qamar HY, Ali Q, Naeem H, Riaz M, Amin S, et al. Role of molecular biology in cancer treatment: A review article. Iran J Public Health 2017;46:1475-85. [Google Scholar] |
| 7. | Pollack JR. Cancer genomics. In: The Molecular Basis of Human Cancer. 2016;241:375-91. [Google Scholar] |
| 8. | Balogh EP, Ganz PA, Murphy SB, Nass SJ, Ferrell BR, Stovall E. Patient-centered cancer treatment planning: Improving the quality of oncology care. Summary of an Institute of Medicine workshop. Oncologist 2011;16:1800-5. [Google Scholar] |
| 9. | Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153:17-37. [Google Scholar] |
| 10. | Collisson EA, Cho RJ, Gray JW. What are we learning from the cancer genome? Nat Rev Clin Oncol 2012;9:621-30. [Google Scholar] |
| 11. | Srivastava S, Grizzle WE. Biomarkers and the genetics of early neoplastic lesions. In: Translational Pathology of Early Cancer; 2012. p. 41-64. [Google Scholar] |
| 12. | Nagpal M, Singh S, Singh P, Chauhan P, Zaidi MA. Tumor markers: A diagnostic tool. Natl J Maxillofac Surg 2016;7:17-20. [Google Scholar] |
| 13. | Irons RD, Stillman WS. The process of leukemogenesis. Environ Health Perspect 1996;104 Suppl 6:1239-46. [Google Scholar] |
| 14. | Vineis P, Matullo G, Manuguerra M. An evolutionary paradigm for carcinogenesis? J Epidemiol Community Health 2003;57:89-95. [Google Scholar] |
| 15. | Lichtenstein AV. On evolutionary origin of cancer. Cancer Cell Int 2005;5:5. [Google Scholar] |
| 16. | Peer D. Precision medicine – Delivering the goods? Cancer Lett 2014;352:2-3. [Google Scholar] |
| 17. | Heckman-Stoddard BM, Smith JJ. Precision medicine clinical trials: Defining new treatment strategies. Semin Oncol Nurs 2014;30:109-16. [Google Scholar] |
| 18. | Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, et al. Next-generation sequencing in oncology: Genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 2017;18:308. [Google Scholar] |
| 19. | Yip S, Christofides A, Banerji S, Downes MR, Izevbaye I, Lo B, et al. A Canadian guideline on the use of next-generation sequencing in oncology. Curr Oncol 2019;26:e241-54. [Google Scholar] |
| 20. | Pang LY, Argyle DJ. Veterinary oncology: Biology, big data and precision medicine. Vet J 2016;213:38-45. [Google Scholar] |
| 21. | Le Tourneau C, Borcoman E, Kamal M. Molecular profiling in precision medicine oncology. Nat Med 2019;25:711-2. [Google Scholar] |
| 22. | Kalachand RD, Hennessy BT, Bast RC, Mills GB. Molecular diagnostics in cancer. In: Holland-Frei Cancer Medicine. John Wiley & Sons, 2017. [Google Scholar] |
| 23. | Singer J, Irmisch A, Ruscheweyh HJ, Singer F, Toussaint NC, Levesque MP, et al. Bioinformatics for precision oncology. Brief Bioinform 2019;20:778-88. [Google Scholar] |
| 24. | Kulasingam V, Prassas I, Diamandis EP. Towards personalized tumor markers. NPJ Precis Oncol 2017;1:17. [Google Scholar] |
| 25. | Biankin AV, Piantadosi S, Hollingsworth SJ. Patient-centric trials for therapeutic development in precision oncology. Nature 2015;526:361-70. [Google Scholar] |
| 26. | The Lancet Oncology. Making precision oncology the standard of care. Lancet Oncol 2017;18:835. [Google Scholar] |
| 27. | Mendelsohn J. Personalizing oncology: Perspectives and prospects. J Clin Oncol 2013;31:1904-11. [Google Scholar] |
| 28. | Lichtenstein AV. Cancer: Shift of the paradigm. Med Hypotheses 2008;71:839-50. [Google Scholar] |
| 29. | Golemis EA, Scheet P, Beck TN, Scolnick EM, Hunter DJ, Hawk E, et al. Molecular mechanisms of the preventable causes of cancer in the United States. Genes Dev 2018;32:868-902. [Google Scholar] |
| 30. | Senft D, Leiserson MD, Ruppin E, Ronai ZA. Precision oncology: The road ahead. Trends Mol Med 2017;23:874-98. [Google Scholar] |
| 31. | Brooks JD. Translational genomics: The challenge of developing cancer biomarkers. Genome Res 2012;22:183-7. [Google Scholar] |
| 32. | Kerick M, Isau M, Timmermann B, Sültmann H, Herwig R, Krobitsch S, et al. Targeted high throughput sequencing in clinical cancer settings: Formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics 2011;4:68. [Google Scholar] |
| 33. | Hühns M, Holzmann C, Prall F. Cancer in a bang: Panel next-generation gene sequencing and OncoScan array analysis of a minute colorectal adenocarcinoma and its precursor adenoma. Histopathology 2019;75:605-8. [Google Scholar] |
| 34. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Google Scholar] |
| 35. | Sen N, Cross AM, Lorenzi PL, Khan J, Gryder BE, Kim S, et al. EWS-FLI1 reprograms the metabolism of Ewing sarcoma cells via positive regulation of glutamine import and serine-glycine biosynthesis. Mol Carcinog 2018;57:1342-57. [Google Scholar] |
| 36. | Tu J, Huo Z, Gingold J, Zhao R, Shen J, Lee DF. The histogenesis of Ewing sarcoma. Cancer Rep Rev 2017;1:10.157611. [Google Scholar] |
| 37. | Wakai T, Prasoon P, Hirose Y, Shimada Y, Ichikawa H, Nagahashi M. Next-generation sequencing-based clinical sequencing: Toward precision medicine in solid tumors. Int J Clin Oncol 2019;24:115-22. [Google Scholar] |
| 38. | Nagahashi M, Shimada Y, Ichikawa H, Kameyama H, Takabe K, Okuda S, et al. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci 2019;110:6-15. [Google Scholar] |
| 39. | Carmagnani Pestana R, Groisberg R, Roszik J, Subbiah V. Precision oncology in sarcomas: Divide and conquer. JCO Precis Oncol 2019;3:1-16. [Google Scholar] |
Fulltext Views
1,782
PDF downloads
267