Translate this page into:
The role of melatonin receptor genes and estrogen receptor genes in the pathogenesis of adolescent idiopathic scoliosis: A systematic review
Corresponding Author:
Sarah A Basindwah
Department of Orthopedics, Faculty of Medicine, Umm Al-Qura University, Makkah Almukkaramah, Makkah
Saudi Arabia
sarah.basindwah@gmail.com
| How to cite this article: Basindwah SA, AlHazmi BF, Haji AK, Eid TM, Fallatah SM. The role of melatonin receptor genes and estrogen receptor genes in the pathogenesis of adolescent idiopathic scoliosis: A systematic review. J Musculoskelet Surg Res 2018;2:8-15 |
Abstract
Objective: Adolescent idiopathic scoliosis (AIS) is the most common spinal deformity affecting healthy children. Genetic factors are thought to play a role in the etiology of this condition, along with other factors. The occurrence of the condition in twins, its early onset, and its familial predilection all support the possibility of an underlying genetic cause. This article aims to review the published studies that looked at the association of estrogen and melatonin receptor (ESR and MTNR) genes with the pathogenesis of AIS, to serve as a guide for future work and opportunities in this field. Methods: The following databases were searched: Cochrane, PubMed, Medline, Ovid, ProQuest, and ScienceDirect using relative keywords. The analysis included 14 studies. Results: Of 312 identified studies, only 14 studies (nine for ESR gene and five for MTNR gene) met our inclusion criteria. Only one study found an association between AIS susceptibility and xbal polymorphism on ESR1 gene. MTNR genes were found to be associated with AIS occurrence in large populations or when synergizing with other gene single-nucleotide polymorphisms. MTNR gene studies showed no relation with curve severity, and none of them considered curve progression, whereas five ESR gene studies considered curve progression and severity. Due to the different gene loci examined in various studies, pooled analysis of the results was not possible. Conclusion: We reviewed the genetic association of ESR and MTNR genes. Several studies found supporting evidence for both genes and their association with AIS, despite conflicting results. Further studies on different genes and different ethnic backgrounds are needed.Introduction
Significant advances have been made in the past decades in the field of medicine in general and in identifying the genes responsible for particular diseases, which have led to specific therapy targeted at the affected gene(s). The same approach was used in the field of orthopedics, for example, in osteoarthritis, a disease that affects a large number of elderly population worldwide, where microRNAs are believed to play a role in chondrogenesis and osteoarthritis pathogenesis.[1] Another condition that affects millions of people; degenerative disc disease and various genes were studied and were found to have single-nucleotide polymorphism (SNPs) supporting the idea that up to 75% of intervertebral disc degeneration is thought to be genetically related.[2] Identifying the genes responsible for a particular disease forms the basis for gene therapy. Many studies have been published on the pathogenesis of adolescent idiopathic scoliosis (AIS), a relatively common condition, affecting 2%–4% of adolescents,[3] with a higher prevalence in females,[4] but its main cause remains unknown. There are many theories concerning the pathogenesis of AIS; these include genetic predisposition,[5],[6],[7] abnormal growth, hormonal disturbance, biomechanical factors,[8],[9] and developmental neuromuscular dysfunctions.[10] Although one factor cannot explain all the clinical characteristics of AIS, the genetic factor seems to play a significant role in its pathogenesis.
Several studies also suggested that the causation of AIS is multifactorial and is strongly associated with genetics as it can be found in different members of the same family.[11],[12] A recent Swedish study of twins estimated that overall genetic effects accounted for 38% of the observed phenotypic variance.[13] Of testing 30 candidate genes, 18 unique loci were identified suggesting that AIS might be due to genes segregating in populations.[14] Although not yet confirmed, it is suggested that AIS is a complex genetic trait influenced by multiple predisposing factors.[15] Of the multiple factors that play a role in the pathogenesis of AIS, the genetic factors are the most widely studied with conflicting results.
Various genes were studied to locate the gene(s) with the strongest association with AIS, which will later be of great value to identify those at risk of developing the disease or maybe developing a therapy for AIS at the genetic level.[16],[17] Estrogen receptor (ESR) genes and melatonin receptor (MTNR) genes are among the most studied genes in the pathogenesis of AIS with variable results. The purpose of this systematic review is to look at the association of ESR and MTNR genes with the pathogenesis of AIS, emphasizing the strength, weakness, and results of each publication and to serve as a guide for future work and opportunities in this field. In addition to being two of the most extensively studied genes, we choose ESR and MTNR genes because curve pathogenesis in AIS patients coincides with growth and adolescence.[14] In females, the high estrogen produced in early puberty is extremely important in regulating the growth of the skeleton and maintaining the mass and strength of bone.[18]ESR gene has been shown to be expressed in both human osteoblasts and osteoclasts, and mutations of the ESR1 gene were shown to reduce bone density and delay skeletal growth in affected humans.[12],[19],[20],[21],[22],[23],[24],[25],[26] Moreover, the female-to-male ratio ranges from 1.5:1 to 3:1 and increases substantially with increasing age.[27] The female-to-male ratio rises with the severity of the curve from 1.4:1 in curves from 10° to 20° up to 7.2:1 in curves >40°.[27] This supports the hypothesis that any mutation affecting the pathway modulating estrogen effect can lead to disturbance in bone growth and maturation. This hypothesis has driven many to study the association of SNPs in the ESR genes and AIS etiology.
In an experimental pinealectomy by Thillard in 1959 on newborn chicken, chicken with low melatonin levels developed a spinal deformity similar to scoliosis in humans.[28] This has led many studies to investigate melatonin levels in AIS patients.[29] However, no significant difference in melatonin levels was found between patients and controls.[30] This suggested that AIS in humans may be caused by another component of the melatonin signaling pathway. Theories suggesting melatonin dysfunction as a cause for abnormal skeletal growth were tested,[31] along with the MTNR gene as candidates for AIS etiology.
We aim to review the published studies that looked at the association of ESR and MTNR genes with the pathogenesis of AIS, emphasizing the strength, weakness, and results of each publication and to serve as a guide for future work and opportunities in this field.
Materials and Methods
This systematic review was confirmed to the Preferred Reporting Items for Systematic Review and Meta-Analyses guidelines.
Search strategy
Cochrane, PubMed, Medline, Ovid, ProQuest, and ScienceDirect databases were searched for case–control studies that examined the association between ESR gene or MTNR gene and AIS. Published articles from 1966 up to February 19, 2017, using the following keyword search string: (Adolescent idiopathic scoliosis OR familial idiopathic scoliosis) AND (estrogen receptor gene OR melatonin receptor gene) were identified. Reference lists of included studies were inspected for additional relevant studies. Furthermore, language restriction was applied (only English language), and no publication date restrictions were applied.
Study selection
Search results were screened independently by four reviewers. Disagreements between reviewers were resolved by discussion until consensus was reached. Level 1 screening consisted of evaluating all available information returned by the electronic search (e.g., abstract, title, and keywords). Level 2 screening consisted of evaluating full-text reports for studies deemed potentially eligible after Level 1 screening or for which insufficient information was available to determine eligibility (e.g., no abstract).
Inclusion and exclusion criteria
The selected studies met the following inclusion criteria: (1) case–control study; (2) English language; (3) AIS diagnosed based on clinical and/or radiological examination; and (4) investigated and reported an association between ESR gene or MTNR gene and AIS. The following are the exclusion criteria: (1) reviews or case reports that were not case–control studies; (2) articles written in a language other than the English language; (3) articles looking at signaling pathway or not directly studying the gene receptors; and (4) duplicate publications.
Methodological quality assessment
The methodological quality of the studies was assessed independently by two reviewers, using a modified version of the Newcastle-Ottawa Scale (NOS) for observational studies (case–control). Disagreements between reviewers were resolved by discussion until consensus was reached. Studies with a score of 5 or higher were deemed satisfactory and considered of high methodological quality.
Results
Study inclusion and characteristics
[Figure - 1] provides a summary of the study identification and selection process. A total of 312 studies were identified. After scanning the titles and abstracts, 27 full-text articles were assessed for eligibility. Of those, only 14 studies with a total of 10,187 participants (5344 patients/4843 controls) met the inclusion criteria. The characteristics of nine studies examining the association between different loci of the ESR genes and AIS are shown in [Table - 1]. Four studies were conducted in Chinese populations, two in Japanese populations, two in Caucasian populations, and one in an Italian population. Six of the nine studies recruited only female cases and controls, while the other three studied recruited cases at a female-to-male ratio of approximately 7.5:1.
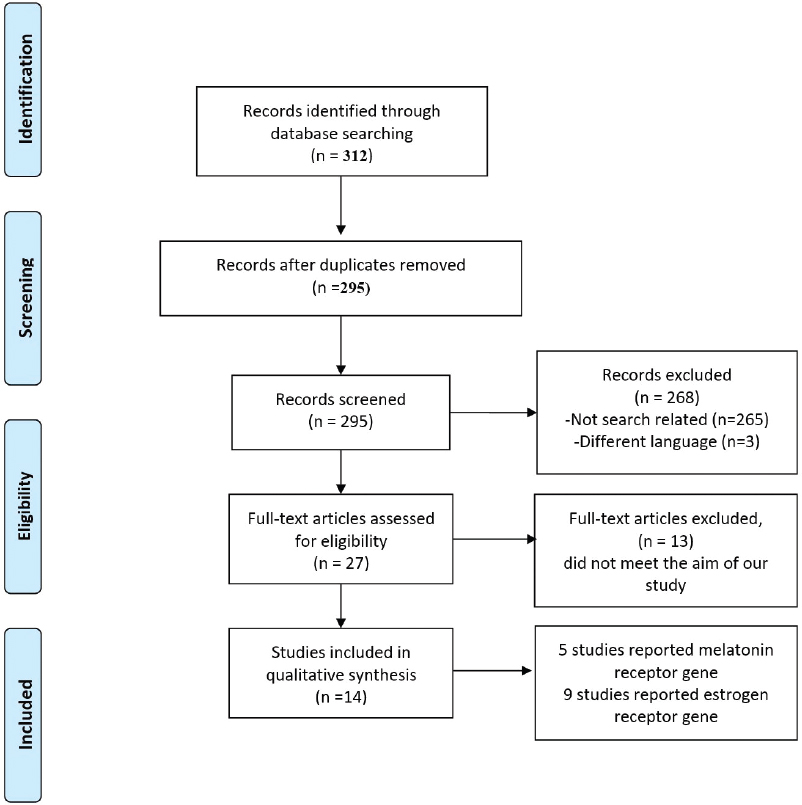 |
| Figure 1: Flow diagram of the study identification and selection process |

[Table - 2] shows the characteristics of five studies examining the association of different loci of the MTNR genes and AIS. Two studies were conducted in Chinese populations, another two were conducted in Caucasian populations, and one study was conducted in a Japanese population. Four of the five studies recruited only female cases. One study recruited 47 families with a total 177 individuals (113 affected/64 not affected). Reporting quality of the included studies was comparatively distinct, but sufficient information was presented to allow for proper assessment of the methodology. All studies assessed using the NOS were of excellent methodological quality, apart from some concerns.
Predisposition
Of the nine studies that looked into the association of ESR genes polymorphisms and AIS, five studies thought of ESR gene as a predisposing factor to AIS.
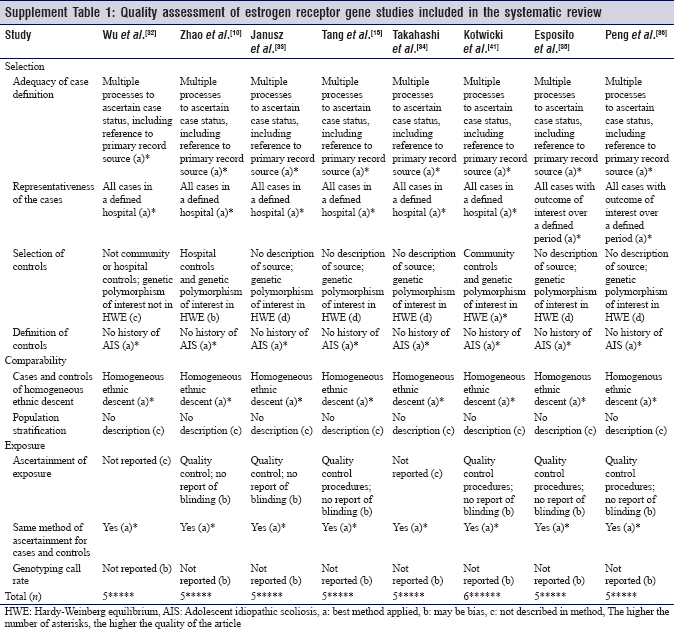
In a Chinese study by Wu et al., significant polymorphism in xbal was found in AIS patients compared to controls.[37]
Both Janusz et al. and Takahashi et al. found no association between the polymorphism of xbaI and the incidence of AIS.[35],[36] Furthermore, Esposito et al. considered xbaI, pstI, stuI, and mseI polymorphisms on ESR1 and did not find >1 mutation per sample.[33]
AlwNI, a locus on ESR2 gene, was thought to play a part of curve predisposition; no association was found in Takahashi et al.'s study.[35]
Peng et al. studied the polymorphism of G-protein coupled receptor gene (GPER), another novel receptor of estrogen, and its association with curve onset; no association was found.[32]
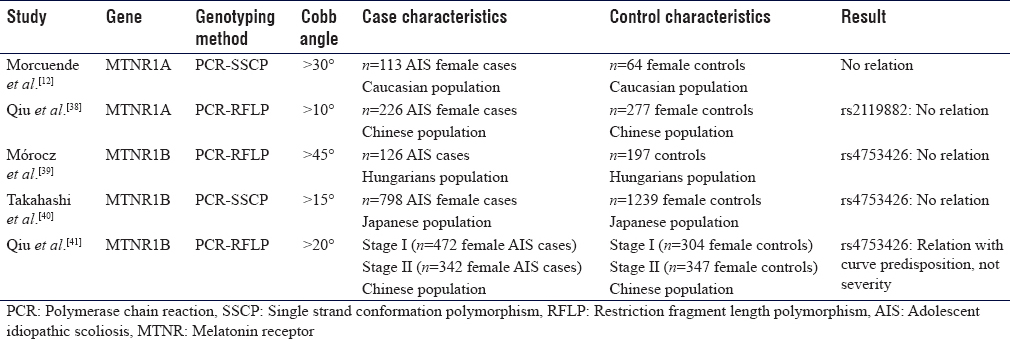
Of the five studies that looked into MTNR genes polymorphisms as a potential factor in the etiology of AIS, three articles studied the polymorphism of rs4753426 in MTNR1B gene and its association with AIS. Morocz et al. studied the MTNR1B gene polymorphism among other genes of other receptors and hormones in Hungarians with AIS, and no association was found between MTNR1B polymorphism and the risk of AIS. However, a combination of leptin (LEP) and MTNR1B gene enhances the susceptibility of AIS.[39] The fact that two or more of predisposing genetic variants of AIS-related factors can be synergistic is foreseeable, giving the complexity of the disease.
A two-stage Chinese study of the same SNP among other four SNPs resulted in a significant association between rs4753426 in MTNR1B gene and predisposition to AIS, after a meta-analysis of the two. Another SNP (rs741837) in the promoter region was also marginally associated with the occurrence AIS.[41]
A replication study on a larger Japanese sample of 798 AIS patients found no significant association between the alleles frequency of the SNP and the predisposition in AIS patients.[40]
Qiu et al. found no association between rs2119882 in MTNR1A gene polymorphism and the occurrence of AIS.[38]
A linkage analysis by Morcuende et al. on 47 Canadian families with AIS found no relation between variant genetic polymorphisms of the MTNR1A gene and the expression of AIS phenotype.[12]
To summarize, only one study found an association between AIS susceptibility and xbal polymorphism on ESR1 gene. Other studies showed no association. MTNR genes were found to be associated with AIS occurrence only in large population studies or when synergizing with other gene polymorphisms.
Severity
Eight of the nine studies about ESR gene polymorphism and its association with AIS considered ESR gene as a factor affecting curve severity.
In 2002, a Japanese study by Inoue et al. found a significant association between the xbaI polymorphism and curve severity but no association in pvuII polymorphisms.[20] Same results were found in 202 Chinese AIS patients in a study by Wu et al.[37] On the other hand, pvuII polymorphism affected curve severity significantly in a study by Zhao et al., but xbaI did not.[10] Janusz et al. and Takashi et al. found no association between curve severity and the polymorphism of xbaI.[35],[36] Similarly, in a larger Chinese study, 540 cases of AIS were studied for SNPs in xbaI and pvuII, and no association was found with curve severity.[15]
Kotwicki et al. found a significant association between AluI (rs4986938) site on ESR2 polymorphism and curve severity but not with AlwNI (rs1256120) and RsaI (rs1256049).[34]
The same AlwNI was thought to play a part in curve severity along with the ESR1 by Takashi's study; no association was found.[35]
Peng et al. found an association between the polymorphism of GPER and curve severity.[32]
Three out of the five articles about MTNR gene polymorphisms studied its association with curve severity in AIS. Qiu et al. and Takahashi et al. found no association between rs2119882 polymorphism on MTNR1A gene and curve severity in AIS patients.[38],[40] A two-stage study by Qiu et al. considered rs4753426 locus among other four loci on the MTNR1B gene. None of the SNPs was related to curve severity in AIS patients.[41] A replication study by Takahashi et al. found no significant association between the alleles frequency of the SNP and curve severity in AIS patients.[40]
To summarize, most of the studies on ESR genes polymorphisms included in this review found an association with curve severity in AIS patients in different loci of the genes. None of the studies of MTNR genes showed association with severity.
Progression
Progression is defined as >5° increase in Cobb angle from the initial assessment.[42]
Five of the estrogen studies concluded that ESR gene polymorphism as a factor affecting curve progression in AIS patients. Inoue et al. found an association between xbal and curve progression, but there was no association with pvull.[20] These results could not be replicated in a study by Tang et al., who found no association between xbaI and pvuII with curve progression.[15] Similarly, both Janusz et al. and Takashi et al. found no association between the xbal polymorphism and curve progression.[35],[36]
A polish study by Kotwicki et al. found a significant association between AluI (rs4986938) site on ESR2 polymorphism and curve progression but not with AlwNI (rs1256120) and RsaI (rs1256049).[34] The same AlwNI was also investigated by Takahashi et al., and no association was found.[35]
None of the studies about MTNR genes polymorphisms considered the progression of AIS.
Discussion
Despite extensive research to know the etiology of AIS, it remains obscure. It is estimated that >30 genes have been studied as candidate genes contributing to susceptibility, severity, and progression of AIS.[14] Among them are the genes responsible for the connective tissue in bone structure. These include genes encoding fibrillin, elastin, collagen I, and collagen II. Studies have shown no association between these genes and AIS.[43],[44],[45] Matrilin, a gene encoding matarlin protein (cartilage matrix protein) involved in the formation of filamentous networks in the extracellular matrices of various tissues, including bone matrix, have been found to be associated with AIS in Italian and Chinese studies.[12],[42],[46] However, a larger Japanese cohort study of (789 cases/1239 controls) found no association.[40]
Matrix metalloproteinase (MMP-3) is an enzyme, which degrades fibronectin, laminin, collagens III, IV, IX, and X, and cartilage proteoglycan. The gene encoding this enzyme and their inhibitors (tissue inhibitors of metalloproteinases) have been studied for the association with AIS. MMP-3 was associated with AIS in a small Italian study.[47] However, neither the association was detected in a larger Chinese study,[48] nor was it directly associated with AIS in a Hungarian sample.[39]
Other sets of genes that were considered are those responsible for bone metabolism, most commonly interleukin 6 (IL-6), leptin (LEP), Bone morphogenetic protein 4 (BMP 4). These genes were studied in a Hungarian sample, along with the MTNR 1B gene. No association was found between independent SNPs and AIS.[39] However, IL-6 was found to be associated with AIS predisposition in a small Italian study.[47] Neither this was confirmed either in a larger Chinese cohort study,[48] nor the previously mentioned Hungarian study.[39] In the same Hungarian sample, Morocz et al. found a synergistic effect of paired SNPs on the risk of AIS formation.
In a small Chinese study, calmodulin 1 receptor gene (CALM1) was associated with the predisposition of a double curve.[49]
In a Japanese study with a sample of 304 females with AIS, Vitamin D receptor (VDR) gene was not associated with the progression of AIS.[20] However, in a Korean study, it was found that VDR is associated with low bone mineral density and double curve formation.[50]
In a case-only Korean study, receptor activator of nuclear factor-κB (RANK), now known as tumor necrosis factor receptor superfamily member 11a (NFKB) activator (TNFRSF11A), as well as a RANK ligand, was not associated with AIS.[51] The same study found an association between osteoprotegerin, now known as tumor necrosis factor receptor superfamily member 11B (TNFRSF11B), and low bone mineral density.
To summarize, studies of genes responsible for connective tissue and bone metabolism that showed association with AIS were small studies that may have been underpowered. Larger studies of the same genes were not able to replicate the same results. Further studies with larger samples and different races are required to determine which genes play a role in the etiology of AIS.
Although proper case selection was done in all the included studies, only five studies selected controls from the community,[34],[38],[39],[40],[41] and other studies either recruited controls from the hospital or did not mention further details about controls. This makes it difficult to assess for selection bias. Population stratification could confuse the genetic associations by making false associations. No population stratification was mentioned in any of the 14 studies [Supplement Table 1] and [Supplement Table 2].
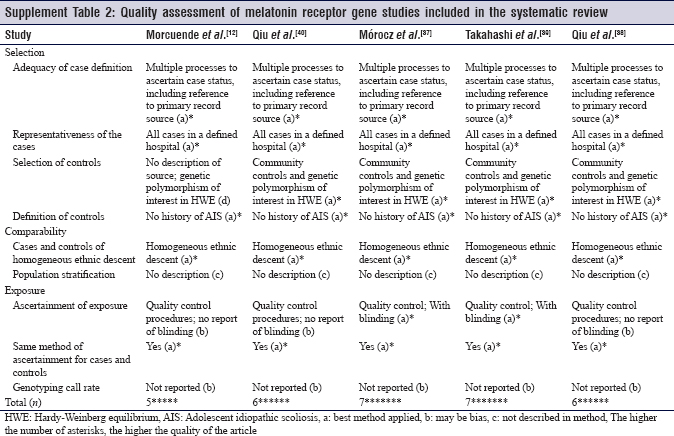
It is apparent that the effort to know the genetic etiology and progression of AIS was distributed toward different directions. Over time, studies were directed towards genes showing more promising results. And even then, researchers have tried to search for different loci in the same gene. A pooled analysis of different studies is only possible if they considered the same locus of a specific gene. Since this review included both ESR genes and MTNR genes, there are a large number of loci that cannot be pooled together. Nonetheless, six out of the nine studies about ESR genes considered xbal as a potential locus for predisposition, severity and progression. Another locus that was considered in five out of the nine ESR genes studies is pvull. Each locus can undergo pooled analysis separately. A systematic review and meta-analysis of the famous xbal and pvull loci on the ESR1 gene was carried by Yang in 2014. The review included five Asian articles [10],[15],[35],[37],[49] and a Polish article [36] concluded no obvious association between xbal and pvull and the risk of developing AIS.[52] Another systematic review was done analyzing four studies about the xbal association to the risk of developing AIS. No association was found.[15],[35],[36],[37] Of note, the xbal could be associated with severity and progression of the curve rather than being a predisposing locus.[53]
Zhao et al. did a systematic review and meta-analysis involving a locus in the ESR2 (rs1256120). Three studies of Chinese,[49] Japanese,[35] and Caucasian [34] populations were analyzed and concluded that the rs1256120 is neither a predisposing factor nor a disease-modifying gene.[54]
As for the MTNR genes associated with AIS, three studies about MTNR1B gene considered rs4753426 as a potential predisposing locus for AIS. A systematic review and meta-analysis carried by Yang et al. included four out of the five MTNR studies found a significant association between a rs4753426 polymorphism in the MTNR1B gene and AIS,[38],[39],[40],[41] especially in the Asian population.[55]
The same locus was considered with another locus from the MTNR1B (rs4753426) in another systematic review of Asian and Caucasian populations in five studies. The study found no obvious association between the two loci and the risk of developing AIS.[56]
Studies included in this review were confined in certain geographical areas. The Chinese population studied in six of the fourteen articles, followed by Caucasians and then Japanese populations. Only one study included Italian population. To our knowledge, there are no studies about ESR genes or MTNR genes association with AIS in Saudi Arabia. Al-Othman et al. recruited 100 AIS girls and their parents and healthy siblings for genetic analysis of three different markers (D19S216, D19S894, and DS1034) on chromosome 19p13.3., and the marker DS1034 was significantly associated with AIS patients and their fathers.[57]
A weakness of the current review is our inability to perform pooled analysis of the results due to the heterogeneity of the studied gene receptors in various studies. We thought despite this weakness, including all relevant studies would help us gather more information about the predisposition, curve severity, and progression with these receptors genes.
Conclusion
The exact cause of AIS remains unknown, despite years of research looking at the multiple potential causative factors. The strong familial predisposition to the condition along with the findings in twin studies favors the role of genetics in the pathogenesis of AIS. Out of the many genes that have been studied, ESR and MTNR genes are two of the most studied genes with the most promising results. In this article, we reviewed the genetic association of ESR and MTNR genes. While ESR genes show more promising results, MTNR studies were insufficient. Moreover, the studies showed inconsistent results. More studies need to be conducted on the two receptors genes, in similar approaches.
Only specific region samples have been studied, most commonly in Southeastern Asia. Larger populations with different ethnic backgrounds should be studies as different results may be obtained. To our knowledge, there were no studies conducted in Saudi Arabia on ESR or MTNR genes. Candidate loci and other loci in these two genes should be studied on Saudi population.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Authors' contributions
SAB designed the study, set inclusion and exclusion criteria and key words, conducted research, reviewed titles and abstracts, helped in reviewing included articles and wrote the initial and final draft. BFA helped in study design, set inclusion and exclusion criteria and key words, conducted research, helped in screening, assessed the methodological quality of the included articles, helped in writing the initial and final draft. AKH conducted research, helped in article screening, assessed the methodological quality of included articles, helped in writing the initial and final draft. TME conducted research, helped in article screening helped in writing the initial and final draft. SMF helped in study design, revised included articles, helped in writing the initial draft, and provided logistic support. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
| 1. | Wu C, Tian B, Qu X, Liu F, Tang T, Qin A, et al. MicroRNAs play a role in chondrogenesis and osteoarthritis (review). Int J Mol Med 2014;34:13-23. [Google Scholar] |
| 2. | Martirosyan NL, Patel AA, Carotenuto A, Kalani MY, Belykh E, Walker CT, et al. Genetic alterations in intervertebral disc disease. Front Surg 2016;3:59. [Google Scholar] |
| 3. | Roach JW. Adolescent idiopathic scoliosis. Orthop Clin North Am 1999;30:353-65, vii-viii. [Google Scholar] |
| 4. | Rogala EJ, Drummond DS, Gurr J. Scoliosis: Incidence and natural history. A prospective epidemiological study. J Bone Joint Surg Am 1978;60:173-6. [Google Scholar] |
| 5. | Kelly PJ, Eisman JA, Sambrook PN. Interaction of genetic and environmental influences on peak bone density. Osteoporos Int 1990;1:56-60. [Google Scholar] |
| 6. | Cowell HR, Hall JN, MacEwen GD. Genetic aspects of idiopathic scoliosis. A Nicholas Andry Award essay, 1970. Clinical Orthopaedics and Related Research 1972;86:121-31. [Google Scholar] |
| 7. | Czeizel A, Bellyei A, Barta O, Magda T, Molnar L. Genetics of adolescent idiopathic scoliosis. J Med Genetics 1978;15:424-7. [Google Scholar] |
| 8. | Andriacchi T, Schultz A, Belytschko T, Galante J. A model for studies of mechanical interactions between the human spine and rib cage. J Biomech 1974;7:497-507. [Google Scholar] |
| 9. | Arkin AM. The mechanism of rotation in combination with lateral deviation in the normal spine. J Bone Joint Surg Am 1950;32:180-8. [Google Scholar] |
| 10. | Zhao D, Qiu GX, Wang YP, Zhang JG, Shen JX, Wu ZH, et al. Association between adolescent idiopathic scoliosis with double curve and polymorphisms of calmodulin1 gene/estrogen receptor-α gene. Orthop Surg 2009;1:222-30. [Google Scholar] |
| 11. | Man GC, Wong JH, Wang WW, Sun GQ, Yeung BH, Ng TB, et al. Abnormal melatonin receptor 1B expression in osteoblasts from girls with adolescent idiopathic scoliosis. J Pineal Res 2011;50:395-402. [Google Scholar] |
| 12. | Morcuende JA, Minhas R, Dolan L, Stevens J, Beck J, Wang K, et al. Allelic variants of human melatonin 1A receptor in patients with familial adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2003;28:2025-8. [Google Scholar] |
| 13. | Grauers A, Rahman I, Gerdhem P. Heritability of scoliosis. Eur Spine J 2012;21:1069-74. [Google Scholar] |
| 14. | Gorman KF, Julien C, Moreau A. The genetic epidemiology of idiopathic scoliosis. Eur Spine J 2012;21:1905-19. [Google Scholar] |
| 15. | Tang NL, Yeung HY, Lee KM, Hung VW, Cheung CS, Ng BK, et al. A relook into the association of the estrogen receptor [alpha] gene (PvuII, xbaI) and adolescent idiopathic scoliosis: A study of 540 Chinese cases. Spine (Phila Pa 1976) 2006;31:2463-8. [Google Scholar] |
| 16. | Nada D, Julien C, Samuels ME, Moreau A. A Replication study for association of Lbx1 locus with adolescent idiopathic scoliosis in French-canadian population. Spine 2017. [Google Scholar] |
| 17. | Ryzhkov II, Borzilov EE, Churnosov MI, Ataman AV, Dedkov AA, Polonikov AV, et al. Transforming growth factor beta 1 is a novel susceptibility gene for adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2013;38:E699-704. [Google Scholar] |
| 18. | Miura K, Namba N, Fujiwara M, Ohata Y, Ishida H, Kitaoka T, et al. An overgrowth disorder associated with excessive production of cGMP due to a gain-of-function mutation of the natriuretic peptide receptor 2 gene. PLoS One 2012;7:e42180. [Google Scholar] |
| 19. | Bashiardes S, Veile R, Allen M, Wise CA, Dobbs M, Morcuende JA, et al. SNTG1, the gene encoding gamma1-syntrophin: A candidate gene for idiopathic scoliosis. Hum Genet 2004;115:81-9. [Google Scholar] |
| 20. | Inoue M, Minami S, Nakata Y, Kitahara H, Otsuka Y, Isobe K, et al. Association between estrogen receptor gene polymorphisms and curve severity of idiopathic scoliosis. Spine (Phila Pa 1976) 2002;27:2357-62. [Google Scholar] |
| 21. | Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, Orimo H. Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res 1996;11:306-11. [Google Scholar] |
| 22. | Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 2003;33:177-82. [Google Scholar] |
| 23. | Matsubara Y, Murata M, Kawano K, Zama T, Aoki N, Yoshino H, et al. Genotype distribution of estrogen receptor polymorphisms in men and postmenopausal women from healthy and coronary populations and its relation to serum lipid levels. Arterioscler Thromb Vasc Biol 1997;17:3006-12. [Google Scholar] |
| 24. | Nadeau JH. Modifier genes and protective alleles in humans and mice. Curr Opin Genet Dev 2003;13:290-5. [Google Scholar] |
| 25. | Wang Z, Yoshida S, Negoro K, Kennedy S, Barlow D, Maruo T, et al. Polymorphisms in the estrogen receptor beta gene but not estrogen receptor alpha gene affect the risk of developing endometriosis in a Japanese population. Fertil Steril 2004;81:1650-6. [Google Scholar] |
| 26. | Yamada Y, Ando F, Niino N, Ohta S, Shimokata H. Association of polymorphisms of the estrogen receptor alpha gene with bone mineral density of the femoral neck in elderly Japanese women. J Mol Med (Berl) 2002;80:452-60. [Google Scholar] |
| 27. | Konieczny MR, Senyurt H, Krauspe R. Epidemiology of adolescent idiopathic scoliosis. J Child Orthop 2013;7:3-9. [Google Scholar] |
| 28. | Thillard MJ. Vertebral column deformities following epiphysectomy in the chick. C R Hebd Seances Acad Sci 1959;248:1238-40. [Google Scholar] |
| 29. | Girardo M, Bettini N, Dema E, Cervellati S. The role of melatonin in the pathogenesis of adolescent idiopathic scoliosis (AIS). Eur Spine J 2011;20 Suppl 1:S68-74. [Google Scholar] |
| 30. | Sadat-Ali M, al-Habdan I, al-Othman A. Adolescent idiopathic scoliosis. Is low melatonin a cause? Joint Bone Spine 2000;67:62-4. [Google Scholar] |
| 31. | Zhang HQ, Lu SJ, Tang MX, Chen LQ, Liu SH, Guo CF, et al. Association of estrogen receptor beta gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2009;34:760-4. [Google Scholar] |
| 32. | Peng Y, Liang G, Pei Y, Ye W, Liang A, Su P, et al. Genomic polymorphisms of G-protein estrogen receptor 1 are associated with severity of adolescent idiopathic scoliosis. Int Orthop 2012;36:671-7. [Google Scholar] |
| 33. | Esposito T, Uccello R, Caliendo R, Di Martino GF, Gironi Carnevale UA, Cuomo S, et al. Estrogen receptor polymorphism, estrogen content and idiopathic scoliosis in human: A possible genetic linkage. J Steroid Biochem Mol Biol 2009;116:56-60. [Google Scholar] |
| 34. | Kotwicki T, Janusz P, Andrusiewicz M, Chmielewska M, Kotwicka M. Estrogen receptor 2 gene polymorphism in idiopathic scoliosis. Spine (Phila Pa 1976) 2014;39:E1599-607. [Google Scholar] |
| 35. | Takahashi Y, Matsumoto M, Karasugi T, Watanabe K, Chiba K, Kawakami N, et al. Replication study of the association between adolescent idiopathic scoliosis and two estrogen receptor genes. J Orthop Res 2011;29:834-7. [Google Scholar] |
| 36. | Janusz P, Kotwicki T, Andrusiewicz M, Kotwicka M. XbaI and PvuII polymorphisms of estrogen receptor 1 gene in females with idiopathic scoliosis: No association with occurrence or clinical form. PLoS One 2013;8:e76806. [Google Scholar] |
| 37. | Wu J, Qiu Y, Zhang L, Sun Q, Qiu X, He Y, et al. Association of estrogen receptor gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2006;31:1131-6. [Google Scholar] |
| 38. | Qiu XS, Tang NL, Yeung HY, Cheng JC, Qiu Y. Lack of association between the promoter polymorphism of the MTNR1A gene and adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2008;33:2204-7. [Google Scholar] |
| 39. | Mórocz M, Czibula A, Grózer ZB, Szécsényi A, Almos PZ, Raskó I, et al. Association study of BMP4, IL6, leptin, MMP3, and MTNR1B gene promoter polymorphisms and adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2011;36:E123-30. [Google Scholar] |
| 40. | Takahashi Y, Matsumoto M, Karasugi T, Watanabe K, Chiba K, Kawakami N, et al. Lack of association between adolescent idiopathic scoliosis and previously reported single nucleotide polymorphisms in MATN1, MTNR1B, TPH1, and IGF1 in a Japanese population. J Orthop Res 2011;29:1055-8. [Google Scholar] |
| 41. | Qiu XS, Tang NL, Yeung HY, Lee KM, Hung VW, Ng BK, et al. Melatonin receptor 1B (MTNR1B) gene polymorphism is associated with the occurrence of adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 2007;32:1748-53. [Google Scholar] |
| 42. | Montanaro L, Parisini P, Greggi T, Di Silvestre M, Campoccia D, Rizzi S, et al. Evidence of a linkage between matrilin-1 gene (MATN1) and idiopathic scoliosis. Scoliosis 2006;1:21. [Google Scholar] |
| 43. | Cao XB, Qiu Y, Qiu XS. FBN3 gene polymorphisms in adolescent idiopathic scoliosis patients. Zhonghua Yi Xue Za Zhi 2008;88:3053-8. [Google Scholar] |
| 44. | Carr AJ, Ogilvie DJ, Wordsworth BP, Priestly LM, Smith R, Sykes B. Segregation of structural collagen genes in adolescent idiopathic scoliosis. Clinical Orthopaedics and Related Research 1992;(274):305-10. [Google Scholar] |
| 45. | Miller NH, Mims B, Child A, Milewicz DM, Sponseller P, Blanton SH, et al. Genetic analysis of structural elastic fiber and collagen genes in familial adolescent idiopathic scoliosis. J Orthop Res 1996;14:994-9. [Google Scholar] |
| 46. | Chen Z, Tang NL, Cao X, Qiao D, Yi L, Cheng JC, et al. Promoter polymorphism of matrilin-1 gene predisposes to adolescent idiopathic scoliosis in a Chinese population. Eur J Hum Genet 2009;17:525-32. [Google Scholar] |
| 47. | Aulisa L, Papaleo P, Pola E, Angelini F, Aulisa AG, Tamburrelli FC, et al. Association between IL-6 and MMP-3 gene polymorphisms and adolescent idiopathic scoliosis: A case-control study. Spine (Phila Pa 1976) 2007;32:2700-2. [Google Scholar] |
| 48. | Liu Z, Tang NL, Cao XB, Liu WJ, Qiu XS, Cheng JC, et al. Lack of association between the promoter polymorphisms of MMP-3 and IL-6 genes and adolescent idiopathic scoliosis: A case-control study in a Chinese Han population. Spine (Phila Pa 1976) 2010;35:1701-5. [Google Scholar] |
| 49. | Zhao D, Qiu GX, Wang YP. Is calmodulin 1 gene/estrogen receptor-alpha gene polymorphisms correlated with double curve pattern of adolescent idiopathic scoliosis? Zhonghua Yi Xue Za Zhi 2008;88:2452-6. [Google Scholar] |
| 50. | Suh KT, Eun IS, Lee JS. Polymorphism in Vitamin D receptor is associated with bone mineral density in patients with adolescent idiopathic scoliosis. Eur Spine J 2010;19:1545-50. [Google Scholar] |
| 51. | Eun IS, Park WW, Suh KT, Kim JI, Lee JS. Association between osteoprotegerin gene polymorphism and bone mineral density in patients with adolescent idiopathic scoliosis. Eur Spine J 2009;18:1936-40. [Google Scholar] |
| 52. | Yang M, Li C, Li M. The estrogen receptor alpha gene (XbaI, PvuII) polymorphisms and susceptibility to idiopathic scoliosis: a meta-analysis. Journal of orthopaedic science : official journal of the Japanese Orthopaedic Association. 2014;19:713-21. [Google Scholar] |
| 53. | Chen S, Zhao L, Roffey DM, Phan P, Wai EK. Association between the ESR1-351A>G single nucleotide polymorphism (rs9340799) and adolescent idiopathic scoliosis: A systematic review and meta-analysis. Eur Spine J 2014;23:2586-93. [Google Scholar] |
| 54. | Zhao L, Roffey DM, Chen S. Association between the estrogen receptor beta (ESR2) rs1256120 single nucleotide polymorphism and adolescent idiopathic scoliosis: A systematic review and meta-analysis. Spine (Phila Pa 1976) 2017;42:871-8. [Google Scholar] |
| 55. | Yang P, Liu H, Lin J, Yang H. The association of rs4753426 polymorphism in the melatonin receptor 1B (MTNR1B) gene and susceptibility to adolescent idiopathic scoliosis: A systematic review and meta-analysis. Pain Physician 2015;18:419-31. [Google Scholar] |
| 56. | Yang M, Wei X, Yang W, Li Y, Ni H, Zhao Y, et al. The polymorphisms of melatonin receptor 1B gene (MTNR1B) (rs4753426 and rs10830963) and susceptibility to adolescent idiopathic scoliosis: A meta-analysis. J Orthop Sci 2015;20:593-600. [Google Scholar] |
| 57. | Al-Othman AA, Sadat-Ali M, Amer AS, Al-Dakheel DA. Genetic markers for adolescent idiopathic scoliosis on chromosome 19p13.3 among Saudi Arabian girls. Asian Spine J 2017;11:167-73. [Google Scholar] |
Fulltext Views
3,317
PDF downloads
928